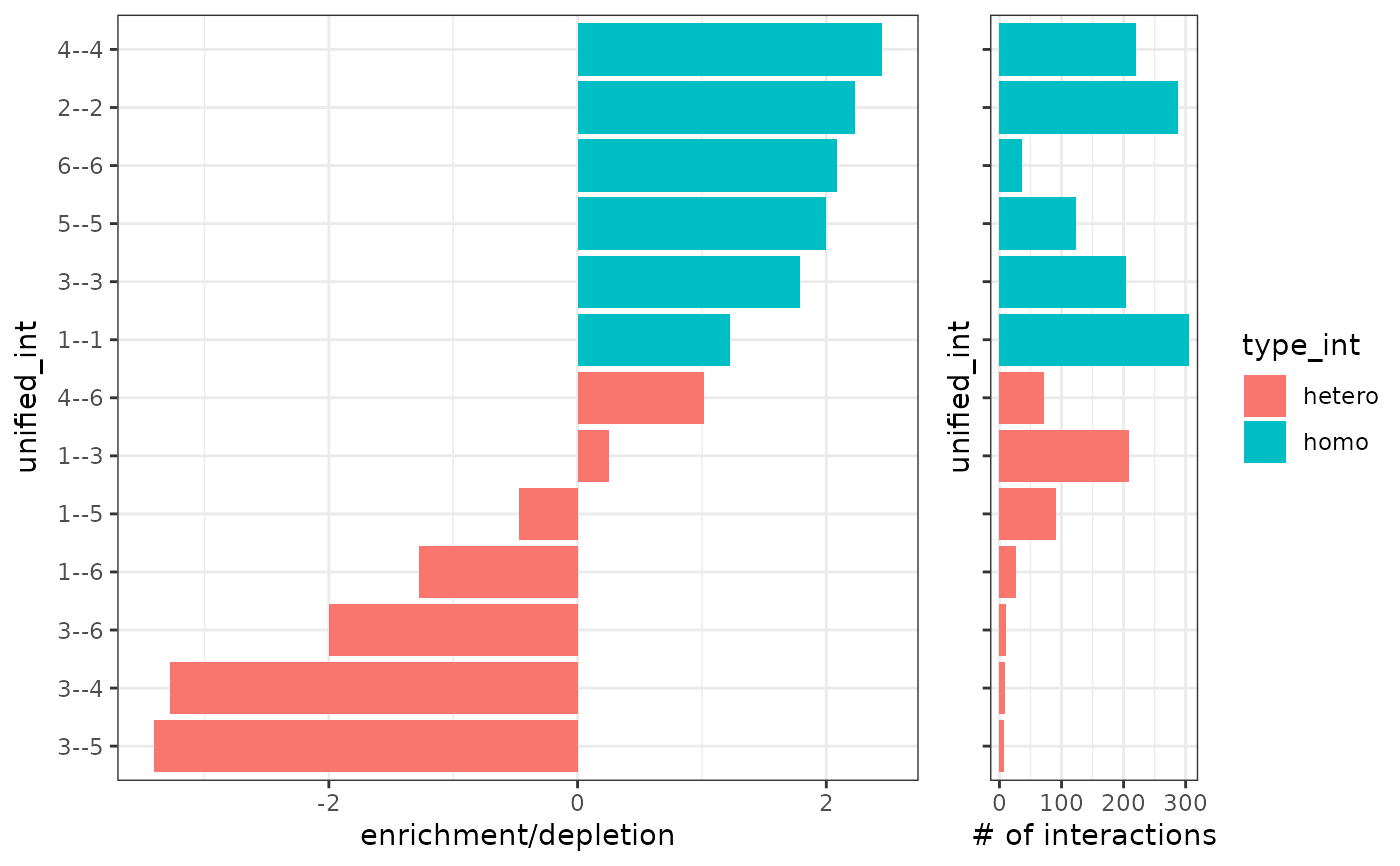
Create barplot from cell-cell proximity scores
cellProximityBarplot(
gobject,
CPscore,
min_orig_ints = 5,
min_sim_ints = 5,
p_val = 0.05,
show_plot = NULL,
return_plot = NULL,
save_plot = NULL,
save_param = list(),
default_save_name = "cellProximityBarplot"
)Arguments
- gobject
giotto object
- CPscore
CPscore, output from cellProximityEnrichment()
- min_orig_ints
filter on minimum original cell-cell interactions
- min_sim_ints
filter on minimum simulated cell-cell interactions
- p_val
p-value
- show_plot
logical. show plot
- return_plot
logical. return ggplot object
- save_plot
logical. save the plot
- save_param
list of saving parameters, see
showSaveParameters- default_save_name
default save name for saving, don't change, change save_name in save_param
Value
ggplot barplot
Details
This function creates a barplot that shows the spatial proximity enrichment or depletion of cell type pairs.
Examples
g <- GiottoData::loadGiottoMini("visium")
#> 1. read Giotto object
#> 2. read Giotto feature information
#> 3. read Giotto spatial information
#> 3.1 read Giotto spatial shape information
#> 3.2 read Giotto spatial centroid information
#> 3.3 read Giotto spatial overlap information
#> 4. read Giotto image information
#> python already initialized in this session
#> active environment : '/usr/bin/python3'
#> python version : 3.12
cellProximityBarplot(
gobject = g,
CPscore = cellProximityEnrichment(g, cluster_column = "leiden_clus")
)