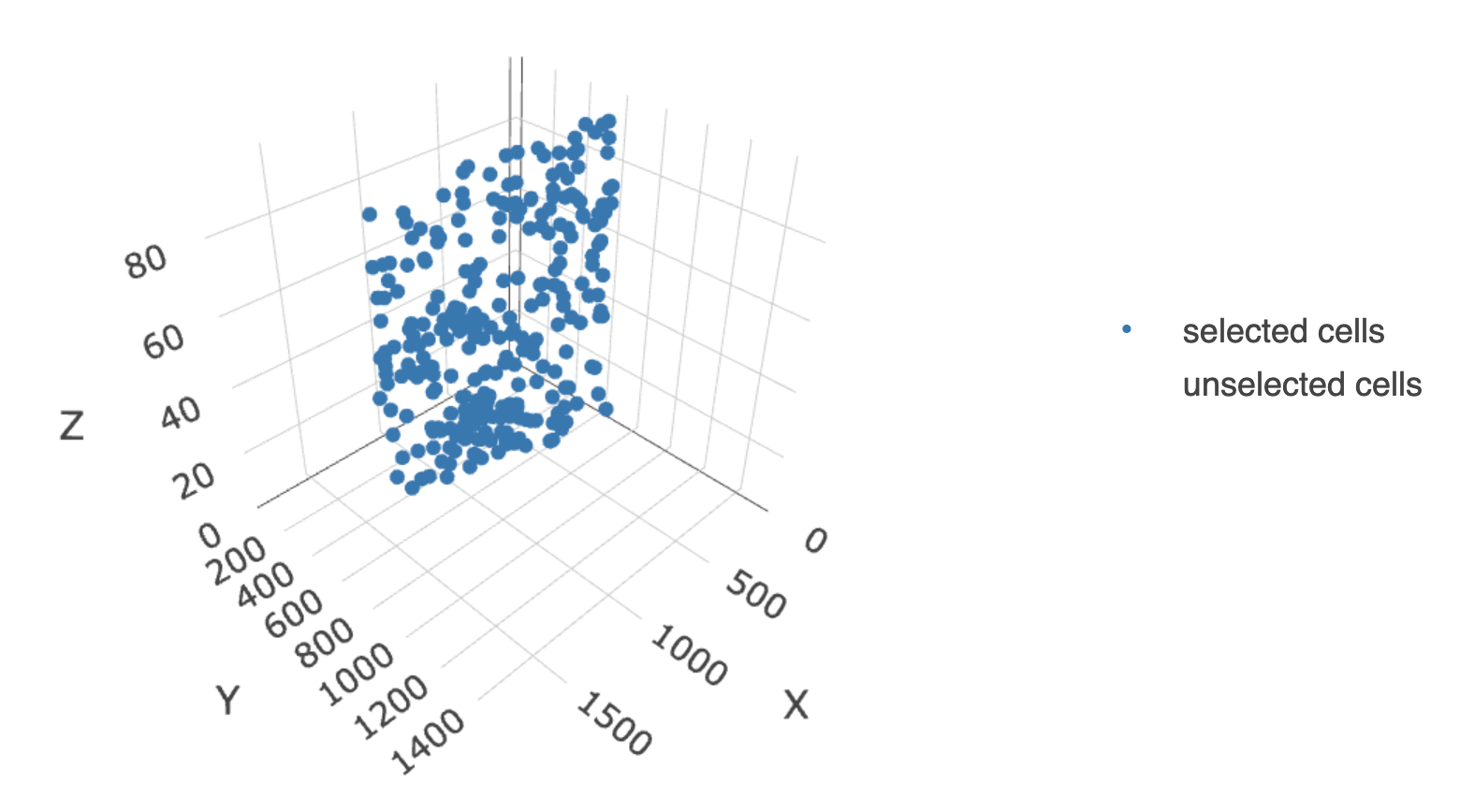
Cross sections in {Giotto} are used to generate an in-silico cross section of a 3D dataset along an arbitrary 2D plane.
A plane may be defined in one of four ways:
"equation" - Provide a vector of 4 numerics to
equation. These will be taken as the A, B, C, D
coefficients in the plane-defining function Ax + By + Cz = D
This method is shown below.
"3 points" - Provide 3D coordinates to
point1, point2, point3 params.
For example:
method = "3 points",
point1 = c(100, 200, 300),
point2 = c(400, 200, 300),
point3 = c(100, 500, 700)"point and norm vector" - Provide a 3D point anchor,
then a 3D vector normal to the plane to set the plane rotation. For
example:
method = "point and norm vector",
point1 = c(100, 200, 300),
normVector = c(0, 1, 0) # Normal points in y direction (horizontal plane)"point and two plane vectors" - Provide a 3D point
anchor and two non-parallel 3D vectors that define the plane rotation.
For example:
method = "point and two plane vectors",
point1 = c(100, 200, 300),
planeVector1 = c(1, 0, 0), # Vector along x-axis
planeVector2 = c(0, 0, 1) # Vector along z-axis1 Setup and load example dataset
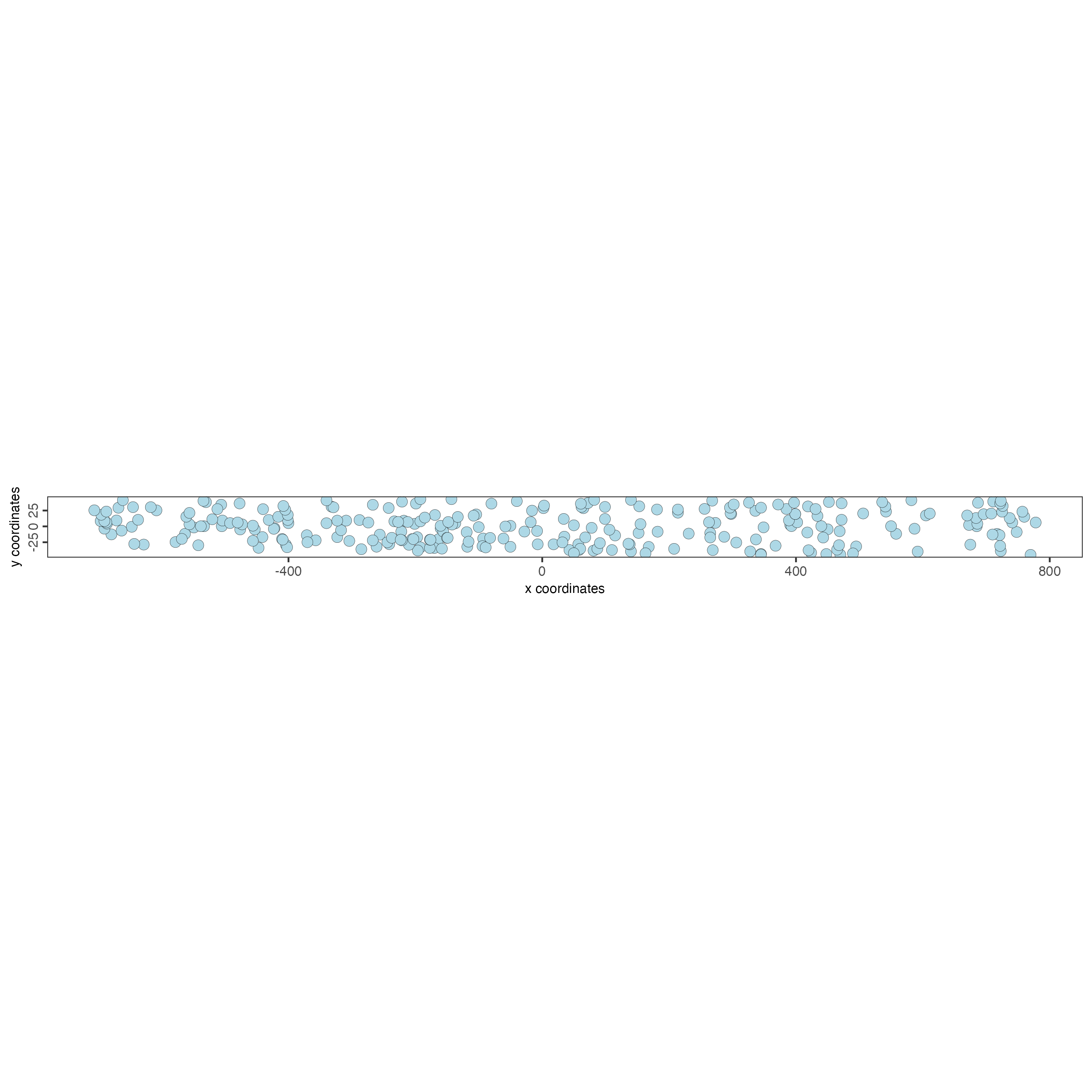
For the example, we use a mini premade giotto object.
This was mini dataset is a randomly sampled subset of a STARmap mouse
brain dataset.
# Ensure Giotto Suite is installed
if(!"Giotto" %in% installed.packages()) {
pak::pkg_install("drieslab/Giotto")
}
# Ensure Giotto Data is installed
if(!"GiottoData" %in% installed.packages()) {
pak::pkg_install("drieslab/GiottoData")
}
library(Giotto)
# load the object
g <- GiottoData::loadGiottoMini("starmap")2 Create Cross Section
Next we define the cross section to create using the
"equation" method.
g <- createCrossSection(g,
method = "equation",
equation = c(0, 1, 0, 600),
extend_ratio = 0.6,
name = "new_cs",
return_gobject = TRUE
)3 Plot cross section
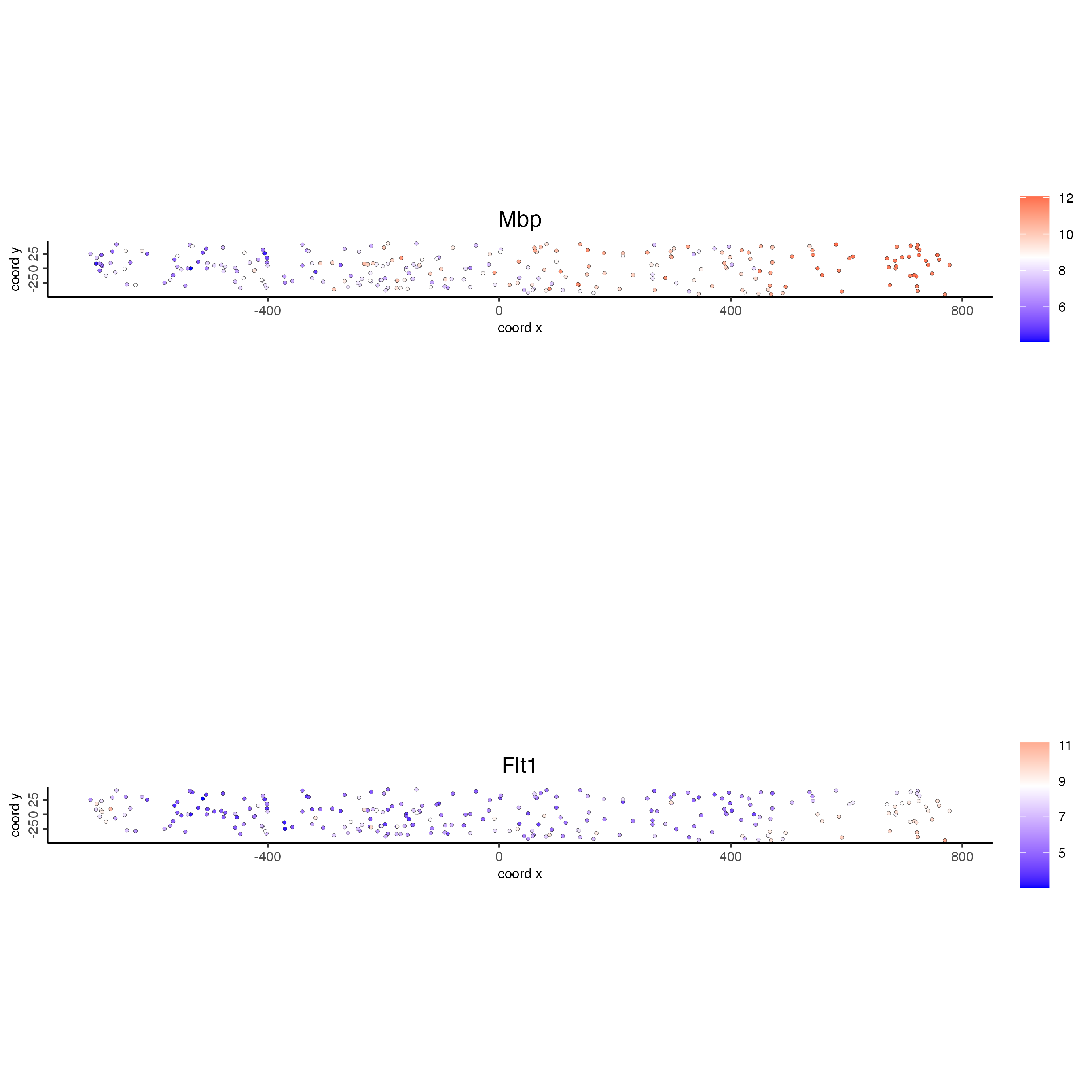
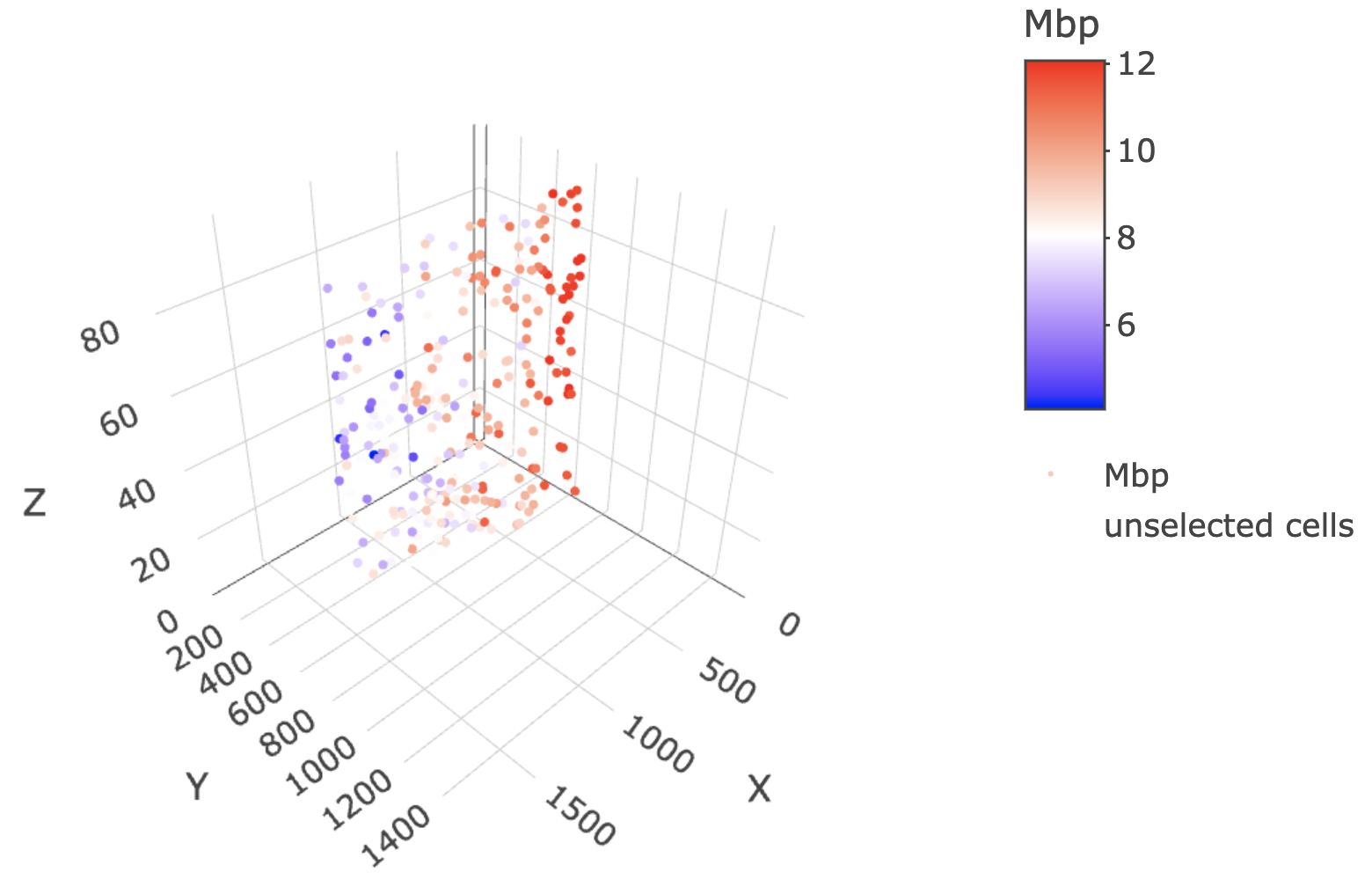
This cross section can then be used in cross section-specific plotting functions.
5 Session Info
R version 4.4.2 (2024-10-31)
Platform: x86_64-apple-darwin20
Running under: macOS Sequoia 15.0.1
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] Giotto_4.1.4 GiottoClass_0.4.3
loaded via a namespace (and not attached):
[1] tidyselect_1.2.1 viridisLite_0.4.2
[3] farver_2.1.2 dplyr_1.1.4
[5] GiottoVisuals_0.2.7 fastmap_1.2.0
[7] SingleCellExperiment_1.26.0 lazyeval_0.2.2
[9] digest_0.6.37 lifecycle_1.0.4
[11] terra_1.7-78 magrittr_2.0.3
[13] compiler_4.4.2 rlang_1.1.4
[15] tools_4.4.2 igraph_2.0.3
[17] utf8_1.2.4 yaml_2.3.10
[19] data.table_1.16.0 knitr_1.48
[21] labeling_0.4.3 S4Arrays_1.4.1
[23] htmlwidgets_1.6.4 reticulate_1.39.0
[25] DelayedArray_0.30.1 abind_1.4-8
[27] withr_3.0.1 purrr_1.0.2
[29] BiocGenerics_0.50.0 grid_4.4.2
[31] stats4_4.4.2 fansi_1.0.6
[33] colorspace_2.1-1 ggplot2_3.5.1
[35] scales_1.3.0 gtools_3.9.5
[37] SummarizedExperiment_1.34.0 cli_3.6.3
[39] rmarkdown_2.28 crayon_1.5.3
[41] ragg_1.3.3 generics_0.1.3
[43] rstudioapi_0.16.0 httr_1.4.7
[45] rjson_0.2.23 zlibbioc_1.50.0
[47] parallel_4.4.2 XVector_0.44.0
[49] matrixStats_1.4.1 vctrs_0.6.5
[51] Matrix_1.7-1 jsonlite_1.8.9
[53] GiottoData_0.2.15 IRanges_2.38.1
[55] S4Vectors_0.42.1 ggrepel_0.9.6
[57] scattermore_1.2 crosstalk_1.2.1
[59] systemfonts_1.1.0 magick_2.8.5
[61] GiottoUtils_0.2.0 plotly_4.10.4
[63] tidyr_1.3.1 glue_1.8.0
[65] codetools_0.2-20 cowplot_1.1.3
[67] gtable_0.3.5 GenomeInfoDb_1.40.1
[69] GenomicRanges_1.56.1 UCSC.utils_1.0.0
[71] munsell_0.5.1 tibble_3.2.1
[73] pillar_1.9.0 htmltools_0.5.8.1
[75] GenomeInfoDbData_1.2.12 R6_2.5.1
[77] textshaping_0.4.0 evaluate_1.0.0
[79] lattice_0.22-6 Biobase_2.64.0
[81] png_0.1-8 backports_1.5.0
[83] SpatialExperiment_1.14.0 Rcpp_1.0.13
[85] SparseArray_1.4.8 checkmate_2.3.2
[87] colorRamp2_0.1.0 xfun_0.47
[89] MatrixGenerics_1.16.0 pkgconfig_2.0.3