Visium spatial transcriptomics does not provide single-cell resolution, making cell type annotation a harder problem. Giotto provides ways to calculate enrichment of specific cell-type signature gene lists.
Parametric Analysis of Gene Set Enrichment (PAGE) and Rank enrichment both aim to determine whether a predefined set of genes show statistically significant differences in expression compared to other genes in the dataset.
1 Setup and load example dataset
# Ensure Giotto Suite is installed
if(!"Giotto" %in% installed.packages()) {
pak::pkg_install("drieslab/Giotto")
}
# Ensure Giotto Data is installed
if(!"GiottoData" %in% installed.packages()) {
pak::pkg_install("drieslab/GiottoData")
}
library(Giotto)
# Ensure the Python environment for Giotto has been installed
genv_exists <- checkGiottoEnvironment()
if(!genv_exists){
# The following command need only be run once to install the Giotto environment
installGiottoEnvironment()
}
# load the object
g <- GiottoData::loadGiottoMini("visium")2 Download the single-cell dataset
GiottoData::getSpatialDataset(dataset = "scRNA_mouse_brain",
directory = "data/scRNA_mouse_brain")3 Create the single-cell object and run the normalization step
results_folder <- "path/to/results"
python_path <- NULL
instructions <- createGiottoInstructions(
save_dir = results_folder,
save_plot = TRUE,
show_plot = FALSE,
python_path = python_path
)
sc_expression <- "data/scRNA_mouse_brain/brain_sc_expression_matrix.txt.gz"
sc_metadata <- data.table::fread("data/scRNA_mouse_brain/brain_sc_metadata.csv")
giotto_SC <- createGiottoObject(expression = sc_expression,
instructions = instructions)
giotto_SC <- addCellMetadata(giotto_SC,
new_metadata = sc_metadata[,2:129])
giotto_SC <- normalizeGiotto(giotto_SC)4 Calculate the cell type markers
markers_scran <- findMarkers_one_vs_all(gobject = giotto_SC,
method = "scran",
expression_values = "normalized",
cluster_column = "Class",
min_feats = 3)
top_markers <- markers_scran[, head(.SD, 10), by = "cluster"]6 Run the enrichment test with PAGE
g <- runPAGEEnrich(gobject = g,
sign_matrix = sign_matrix,
min_overlap_genes = 1)7 Visualize
Create a heatmap showing the enrichment of cell types (from the single-cell data annotation) in the spatial dataset clusters.
cell_types_PAGE <- colnames(sign_matrix)
plotMetaDataCellsHeatmap(gobject = g,
metadata_cols = "leiden_clus",
value_cols = cell_types_PAGE,
spat_enr_names = "PAGE",
x_text_size = 8,
y_text_size = 8)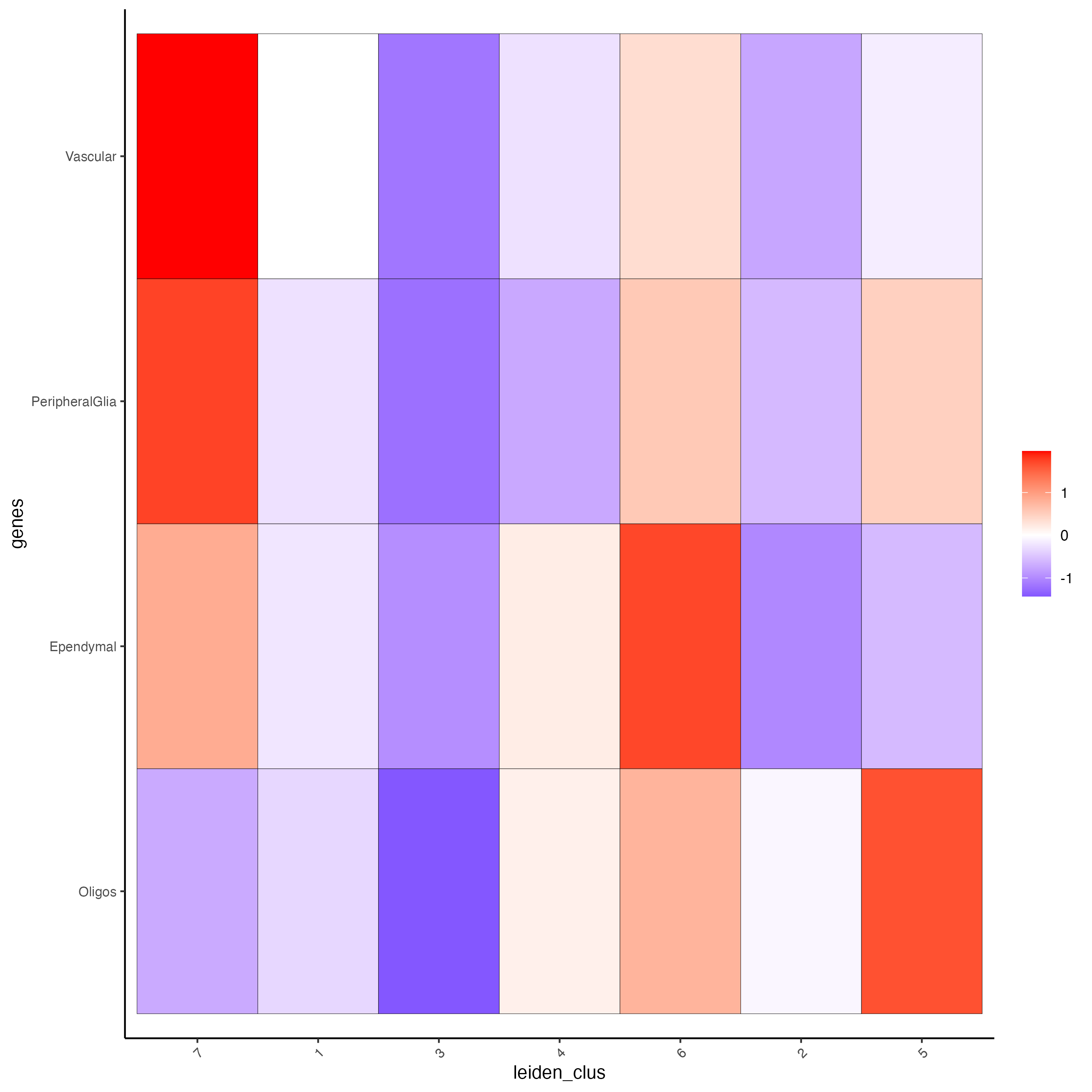
Plot the spatial distribution of the cell types.
spatCellPlot2D(gobject = g,
spat_enr_names = "PAGE",
cell_annotation_values = cell_types_PAGE,
cow_n_col = 2,
coord_fix_ratio = 1,
point_size = 4,
show_legend = TRUE)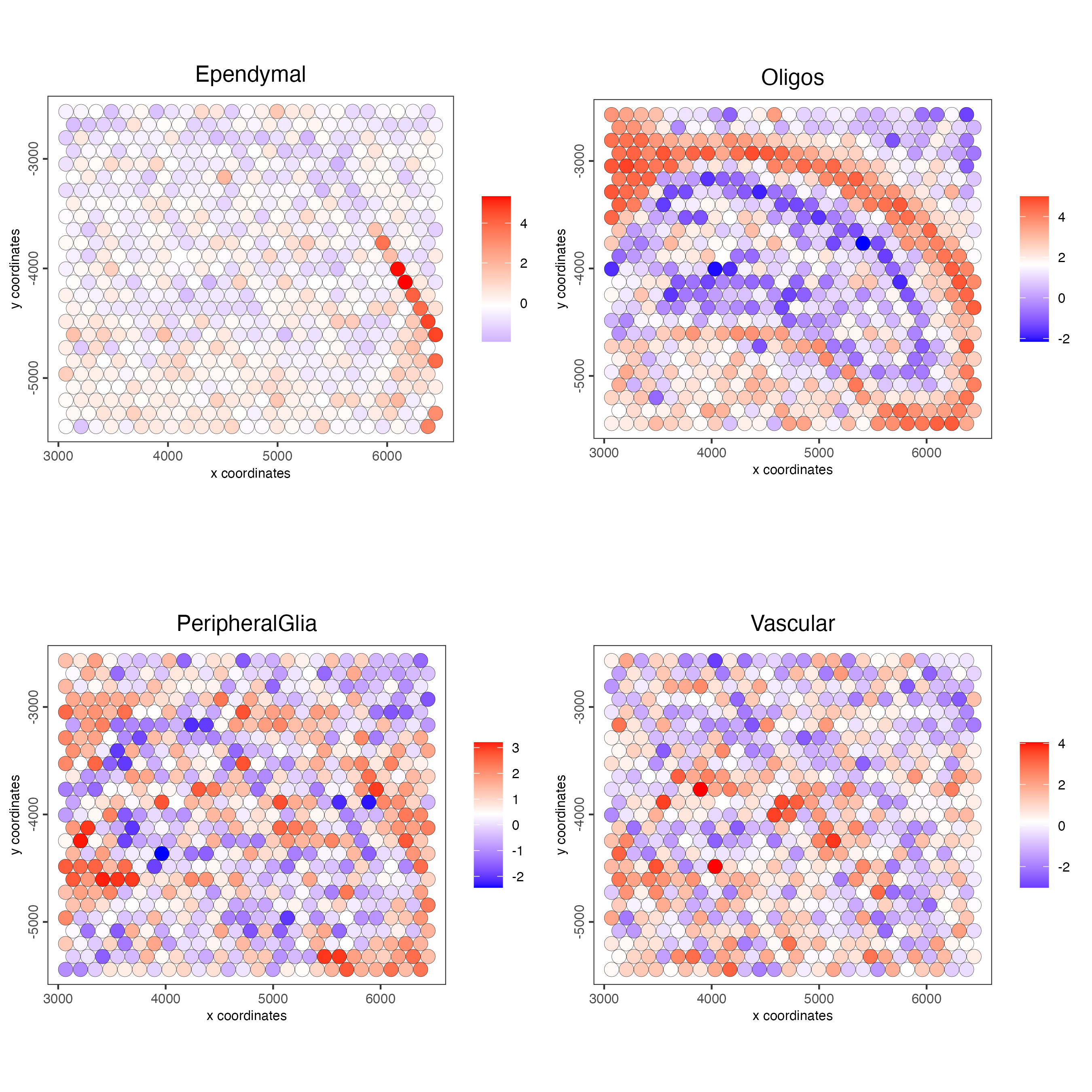
8 Session info
R version 4.4.2 (2024-10-31)
Platform: x86_64-apple-darwin20
Running under: macOS Sequoia 15.0.1
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] Giotto_4.1.4 GiottoClass_0.4.3
loaded via a namespace (and not attached):
[1] colorRamp2_0.1.0 rlang_1.1.4
[3] magrittr_2.0.3 GiottoUtils_0.2.0
[5] matrixStats_1.4.1 compiler_4.4.2
[7] DelayedMatrixStats_1.26.0 systemfonts_1.1.0
[9] png_0.1-8 vctrs_0.6.5
[11] RcppZiggurat_0.1.6 quadprog_1.5-8
[13] pkgconfig_2.0.3 SpatialExperiment_1.14.0
[15] crayon_1.5.3 fastmap_1.2.0
[17] backports_1.5.0 magick_2.8.5
[19] XVector_0.44.0 labeling_0.4.3
[21] scuttle_1.14.0 utf8_1.2.4
[23] rmarkdown_2.28 UCSC.utils_1.0.0
[25] ragg_1.3.3 purrr_1.0.2
[27] Rfast_2.1.0 xfun_0.47
[29] bluster_1.14.0 zlibbioc_1.50.0
[31] beachmat_2.20.0 GenomeInfoDb_1.40.1
[33] jsonlite_1.8.9 DelayedArray_0.30.1
[35] tweenr_2.0.3 BiocParallel_1.38.0
[37] terra_1.7-78 irlba_2.3.5.1
[39] parallel_4.4.2 cluster_2.1.6
[41] R6_2.5.1 RColorBrewer_1.1-3
[43] limma_3.60.4 reticulate_1.39.0
[45] GenomicRanges_1.56.1 scattermore_1.2
[47] Rcpp_1.0.13 SummarizedExperiment_1.34.0
[49] knitr_1.48 R.utils_2.12.3
[51] IRanges_2.38.1 Matrix_1.7-1
[53] igraph_2.0.3 tidyselect_1.2.1
[55] rstudioapi_0.16.0 abind_1.4-8
[57] yaml_2.3.10 codetools_0.2-20
[59] lattice_0.22-6 tibble_3.2.1
[61] Biobase_2.64.0 withr_3.0.1
[63] evaluate_1.0.0 polyclip_1.10-7
[65] scatterpie_0.2.4 RcppParallel_5.1.9
[67] pillar_1.9.0 MatrixGenerics_1.16.0
[69] checkmate_2.3.2 stats4_4.4.2
[71] ggfun_0.1.6 plotly_4.10.4
[73] generics_0.1.3 S4Vectors_0.42.1
[75] ggplot2_3.5.1 sparseMatrixStats_1.16.0
[77] munsell_0.5.1 scales_1.3.0
[79] GiottoData_0.2.15 gtools_3.9.5
[81] glue_1.8.0 metapod_1.12.0
[83] lazyeval_0.2.2 tools_4.4.2
[85] GiottoVisuals_0.2.7 BiocNeighbors_1.22.0
[87] data.table_1.16.0 ScaledMatrix_1.12.0
[89] locfit_1.5-9.10 fs_1.6.4
[91] scran_1.32.0 cowplot_1.1.3
[93] grid_4.4.2 tidyr_1.3.1
[95] edgeR_4.2.1 colorspace_2.1-1
[97] SingleCellExperiment_1.26.0 GenomeInfoDbData_1.2.12
[99] ggforce_0.4.2 BiocSingular_1.20.0
[101] cli_3.6.3 rsvd_1.0.5
[103] textshaping_0.4.0 fansi_1.0.6
[105] S4Arrays_1.4.1 viridisLite_0.4.2
[107] dplyr_1.1.4 gtable_0.3.5
[109] yulab.utils_0.1.7 R.methodsS3_1.8.2
[111] digest_0.6.37 progressr_0.14.0
[113] BiocGenerics_0.50.0 SparseArray_1.4.8
[115] ggrepel_0.9.6 dqrng_0.4.1
[117] farver_2.1.2 rjson_0.2.23
[119] htmlwidgets_1.6.4 htmltools_0.5.8.1
[121] R.oo_1.26.0 lifecycle_1.0.4
[123] httr_1.4.7 statmod_1.5.0
[125] MASS_7.3-61