Split cluster annotations based on a spatial network
Source:R/spatial_clusters.R
spatialSplitCluster.RdSplit cluster annotations based on a spatial network
spatialSplitCluster(
gobject,
spat_unit = NULL,
feat_type = NULL,
spatial_network_name = "Delaunay_network",
cluster_col,
split_clus_name = paste0(cluster_col, "_split"),
missing_id_name = "not_connected",
return_gobject = TRUE
)Arguments
- gobject
giotto object
- spat_unit
spatial unit (e.g. "cell")
- feat_type
feature type (e.g. "rna", "dna", "protein")
- spatial_network_name
character. Name of spatial network to use
- cluster_col
character. Column in metadata containing original clustering
- split_clus_name
character. Name to assign the split cluster results
- return_gobject
logical. Return giotto object
- include_all_ids
logical. Include all ids, including vertex ids not found in the spatial network
Value
giotto object with cluster annotations
Examples
g <- GiottoData::loadGiottoMini("vizgen")
#> 1. read Giotto object
#> 2. read Giotto feature information
#> 3. read Giotto spatial information
#> 3.1 read Giotto spatial shape information
#> 3.2 read Giotto spatial centroid information
#> 3.3 read Giotto spatial overlap information
#> 4. read Giotto image information
#> python already initialized in this session
#> active environment : '/usr/bin/python3'
#> python version : 3.12
activeSpatUnit(g) <- "aggregate"
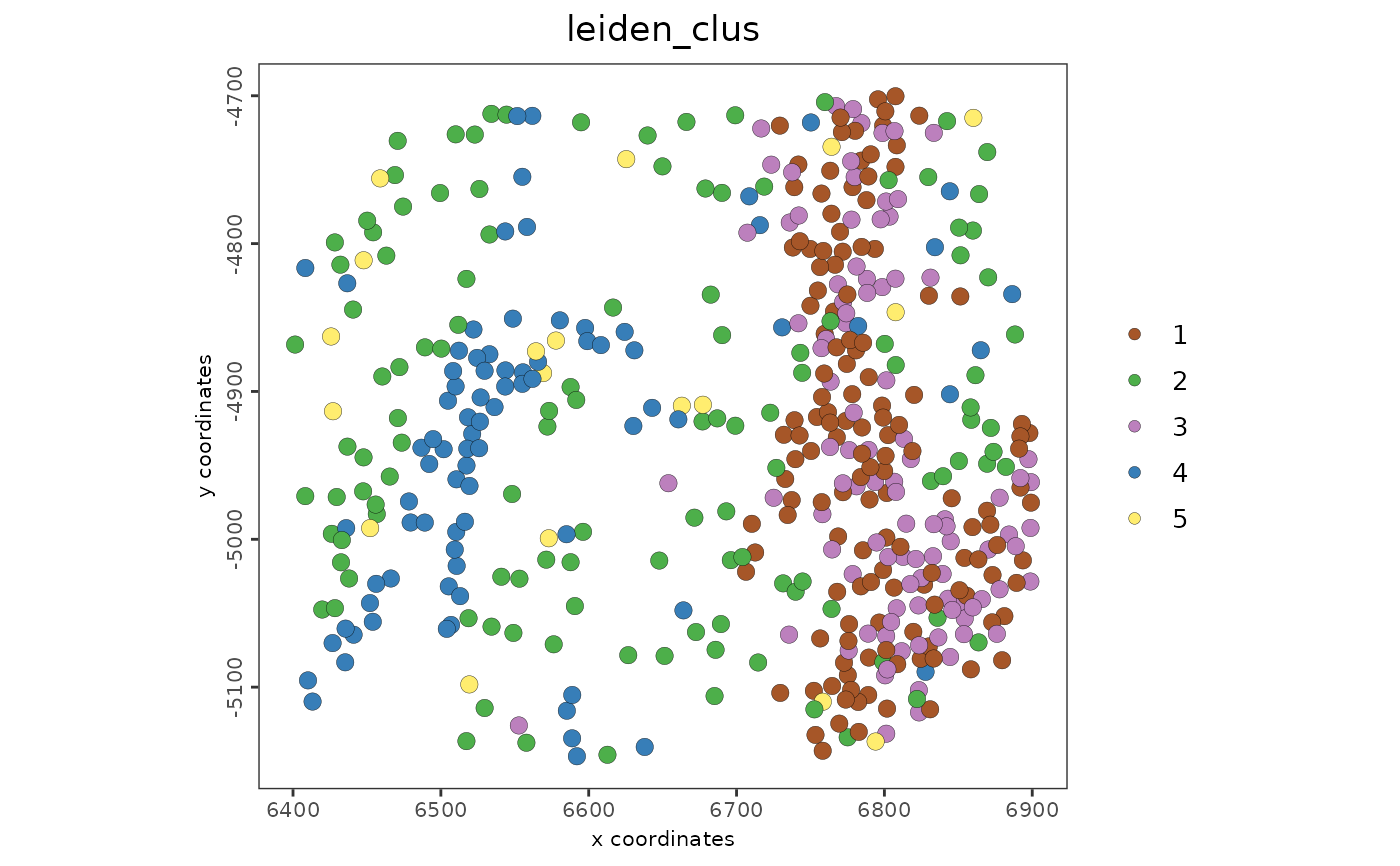
spatPlot2D(g, cell_color = "leiden_clus")
 g <- spatialSplitCluster(g,
cluster_col = "leiden_clus",
split_clus_name = "new"
)
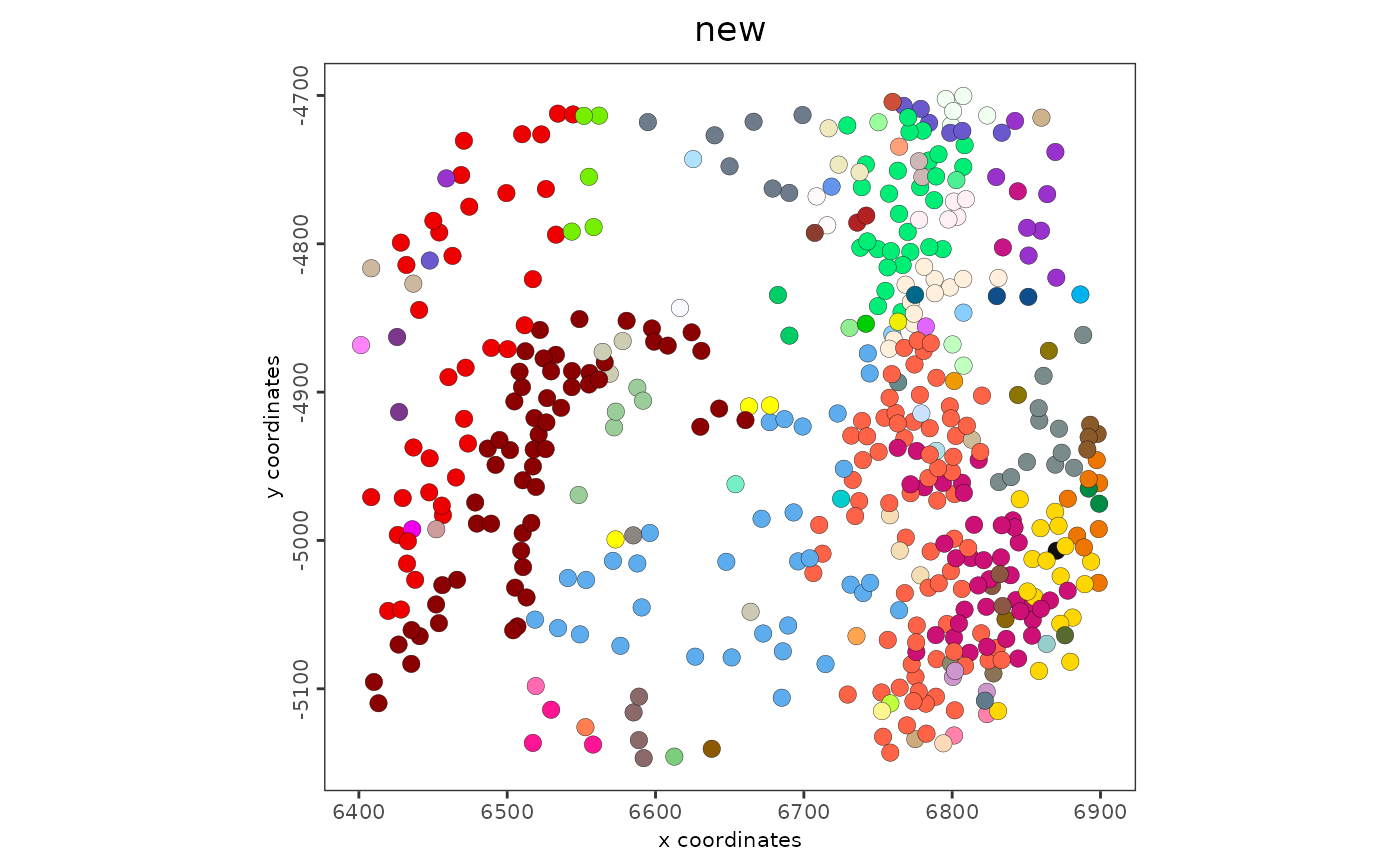
# don't show legend since there are too many categories generated
spatPlot2D(g, cell_color = "new", show_legend = FALSE)
g <- spatialSplitCluster(g,
cluster_col = "leiden_clus",
split_clus_name = "new"
)
# don't show legend since there are too many categories generated
spatPlot2D(g, cell_color = "new", show_legend = FALSE)